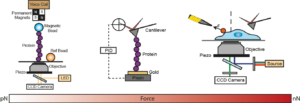
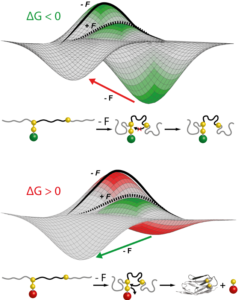
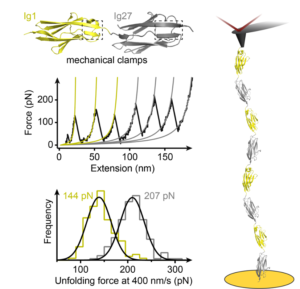
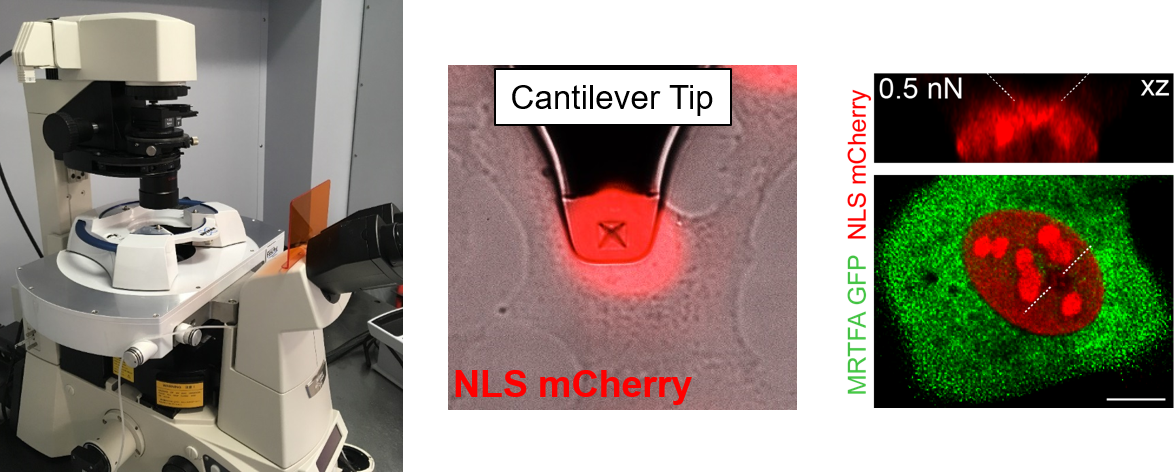
The Garcia-Manyes is interested in mechanobiology at different length- and force-scales. We use a combination of bespoke nanomechanical techniques at the single molecule and at the single cell level to interrogate how mechanical forces affect protein folding, often in the context of cellular mechanotransduction.
1. Mechanochemistry at the single bond level
Using single molecule AFM in the force-clamp mode, we investigate how mechanical forces regulate chemical reactivity in protein substrates. A chemical reaction is classically activated by temperature, light or electricity. Force is an alternative, albeit much less explored, way to promote a chemical reaction. Using a combination of force clamp spectroscopy with molecular engineereing techniques we demonstrated that the SN2 reduction of an individual protein embedded disulfide bond is a force-activated process along a smooth energy landscape. We plan to experimentally reconstruct the full free-energy landscape of a chemical reaction under force. These novel experiments have opened a totally new way of inquiry in the chemistry field.
Key References
- Beedle, A. E*., Mora, M*., Davis, C., Snijders, A., Stirnemann, G., Garcia-Manyes, S. «Forcing the reversibility of a mechanochemical reaction» Nature Communications (2018), 9, 3155.
- Garcia-Manyes, S., Beedle, A.E.M. «Steering chemical reactions with force» Nature Reviews Chemistry (2017)
- Beedle, A. E., Mora, M., Lynham, S., Stirnemann, G., Garcia-Manyes, S. «Tailoring protein nanomechanics with chemical reactivity» Nature Communications (2017), 8, 15658.
- Perales-Calvo, J.; Lezamiz, A.; Garcia-Manyes, S. «The mechanochemistry of a structural zinc finger» The Journal of Physical Chemistry Letters (2015), 6, 3335-3340.
- Beedle, A. E.; Lezamiz, A.; Sitrnemann, G.; Garcia-Manyes, S. «The mechanochemistry of copper reports on the directionality of unfolding in model cupredoxin proteins» Nature Communications (2015), 6, 7894.
2. Mechanobiology at the single protein level – the molecular determinants underpinning mechanical (un)folding
We are interested in studying the nanomechanics of proteins that are in vivo continuously exposed to mechanical forces. Understanding the mechanisms of tissue elasticity requires comprehending how these proteins continuously and reversibly extend and recoil physiologically under mechanical stress. In particular, we want to elucidate the effect of post-translational modifications and chaperone binding on mechanical folding, with direct knock-on effects on protein elasticity.
Key References:
- Perales-Calvo, J., Giganti, D., Stirnemann, G., Garcia-Manyes, S. «The force-dependent mechanisms of DnaK-mediated mechanical folding» Science Advances (2018) 4, 2.
- Beedle, A. E.; Lynham, S.; Garcia-Manyes, S. «Protein S-sulfenylation is a fleeting molecular switch that regulates non-enzymatic oxidative folding» Nature Communications (2016), 7, 12490.
3. Mechanobiology at the single cell level
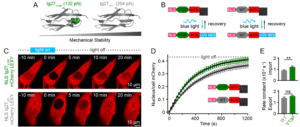
Understanding the molecular mechanisms by which mechanical forces from the extracellular matrix propagate deep inside the cell, and eventually reach the nucleus, is arguably one of the main sough-after question in molecular mechanobiology. In this vein, we have recently discovered that the nuclear pore complex (NPC) regulates the rate of nuclear entry of proteins according to their mechanical properties.
- Infante, E; Stannard, A; Board, S.J.; Rico-Lastres, P.; Panagaki, F.; Beedle, A.E.M.; Sundar Rajan, V.; Rostkova, E.; Lezamiz, A.; Wang, Y.; Gulaidi, S.; Shanahan, C.; Roca-Cusachs, P.; Garcia-Manyes, S. «The mechanical stability of proteins regulates their translocation rate into the cell nucleus», Nature Physics (2019), 15, 973–981.
- Elosegui-Artola, A., Andreu, I., Beedle, A.E.M., Lezamiz, A., Uroz, M., Kosmalska, A., Oria, R., Kechagia, J., Rico-Lastres, P., Le Roux, A., Shanahan, C., Trepat, X., Navajas, D., Garcia-Manyes, S., Roca-Cusachs, P. «Force triggers YAP entry by mechanically regulating transport across nuclear pores» Cell (2017), 171, 1-14.
- Atilla-Gokcumen, G.; Muro, E.; Relat-Goberna, J.; Sasse, S.; Bedigian, A.; Coughlin, M.; Garcia-Manyes, S.; Eggert, U. «Dividing cells regulate their lipid composition and localization» Cell (2014), 3, 428-439.